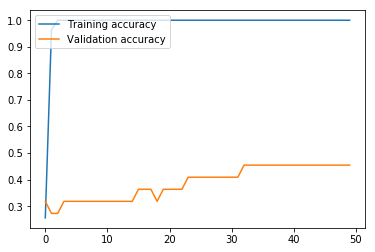
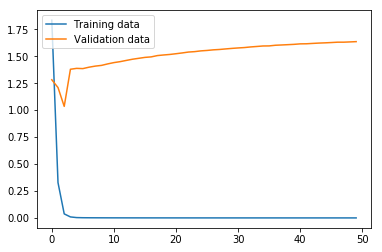
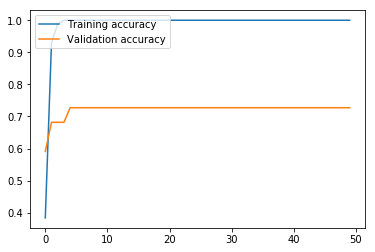
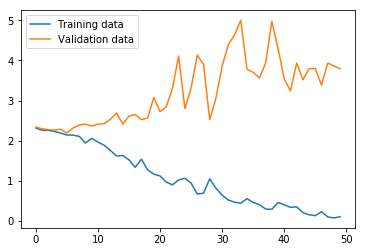
non_nouns_to_exclude = ['puav', 'me', 'hluas', 'kws', 'laus', 'twg', 'uas', \
'laug', 'ub', 'mos2', '22', 'hlob', 'loj', 'coj', '.', ',', \
'ntawd', 'yog', 'tod', 'swb', 'li', 'tuag', '#', 'sau', \
'niag', 'tias', 'lawm', 'ib', 'mos', 'muab', '/', 'muaj', \
'nrog', 'rau', 'luag', 'ua', 'los', 'nws', 'txawm', 'hais', \
'thaum', 'lawv', 'tsi', 'es', 'phem', 'nuav', 'tej', 'has', \
'xav', 'hov', 'kuv', 'ces', 'ntawm', 'tawm', 'lwm', '(', 'kiag',\
'hu', 'cov', 'ntseeg', 'mus', 'ko', 'mas', 'tiag', 'to', \
'yam', 'tag', 'nawb', 'pom', 'miv', 'no', 'peb', 'sib', 'hlub', \
'twb', 'thiab', 'pab', 'leej', 'tsis', '...', 'kawg', 'kom', \
'xwb', 'tau', 'tshiab', 'noj', 'tus', 'qub', 'lub', 'txoj', \
'nyuas', 'thib', 'ntse', 'nyuag', 'thiaj', 'tshab', 'nua', 'koj',\
'tham', 'yau', 'tham', 'saib', 'hauv', 'yees', 'teb', 'luj', \
'txiav', 'tswj', 'xub', 'thaub', 'cuav', 'puas', 'txheeb', 'puag', \
'ruam', 'siab', 'tsim', 'pluag', 'yus', 'tuav', 'rog', 'txawj',\
'mob', 'tub']
partial_words_to_exclude = ['poj', 'tij', 'quas', 'xf', 'dr', 'ntsuj', 'tib', 'tuab', \
'teeb', 'yeeb', 'xeeb', 'kas', 'cawm', 'zuj', 'npau', 'cuj',\
'cwj', 'xov', 'kav', 'kab', 'txheej', 'xib', 'huab', 'pej',\
'phooj']
green_mong_to_exclude = ['mivnyuas', 'nam', 'dlaab', 'puj', 'moob', 'tuabneeg', 'quasyawg',\
'quaspuj', 'dlev', 'tsaj', 'nav', 'qab']
total_proc = [w for w in total_proc if w not in non_nouns_to_exclude]
total_proc = [w for w in total_proc if w not in partial_words_to_exclude]
total_proc = [w for w in total_proc if w not in green_mong_to_exclude]
total_proc = list(set(total_proc))